ADVERTISEMENT
Codon Usage
Database & Resources
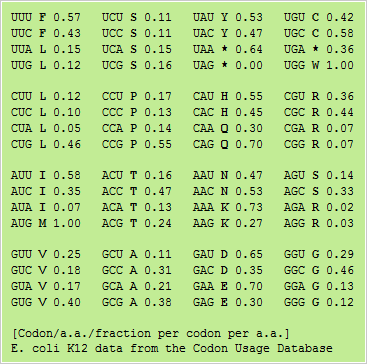
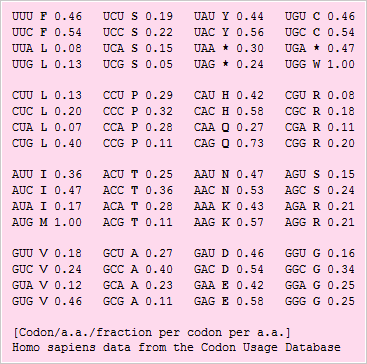
An expanded codon table showing the relative frequence that different codons are used in E. coli genes
Genes are clustered by using factorial correspondence analysis into three classes. Class I contains genes involved in most metabolic processes. Class II genes correspond to genes highly and continuously expressed during exponential growth. Class III genes are implicated in horizontal transfer of DNA.
Codon Analysis
The gcua tool displays the codon quality graphically in two different ways: codon usage frequency and relative adaptiveness values.
This server performs several computations in relation to codon usage and the codon adaptation of DNA or RNA sequences to host organisms.
Codon Optimization
Visual Gene Developer is a specialized gene design software that has many functions to analyze, design, and optimize genes.
OPTIMIZER is an on-line application that optimizes the codon usage of a DNA sequence to increase its expression level.
The Codon Adaptation Tool (JCAT) presents a simple method to adapt the Codon Usage to most sequenced prokaryotic organisms and selected eukaryotic organisms. The codon adaptation plays a major role in cases where foreign genes are expressed in hosts and the codon usage of the host differs from that of the organism where the gene stems from.
This server performs several computations in relation to codon usage and the codon adaptation of DNA or RNA sequences to host organisms.
The Web Bench
The Web Bench
is the essential companion to the biologist, bringing informational resources and a collection of tools & calculators to facilitate work at the bench and analysis of biological data.
Check out the full online bench here
Check out the full online bench here
Sequence Analysis with GenBeans
 Try GenBeans: Best free software for DNA sequence editing!
Try GenBeans: Best free software for DNA sequence editing!
FEEDBACK
Your comments & your suggestionsare appreciated. Please, notify us for resources and tools that you would like to see on this bench!