Summary
- Find your sequences quickly in the Explorer
- No obscure sequence format
- Advanced Sequence Viewer
- Advanced Search Capabilities
- Advanced Restriction Analysis
- Feature Navigator
- Simple & Rapid Primer Design
- Access large sequence files
- GenBeans is free of charge
Find your sequences quickly in the Explorer
No virtual locations or mysterious databases, you add real folders that exist on your computer or on your network and your sequences become immediatly alive:
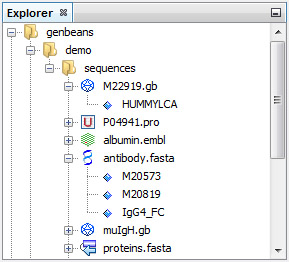
The format of FASTA sequence file is recognized automatically. If GenBeans makes a mistake (e.g. take a polyA for a poly alanine), you change the format and GenBeans will remember on the next session!
No obscure sequence format
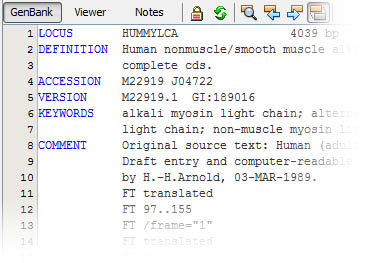
No, GenBeans is not using another of those mysterious, lock-in, binary file formats! GenBeans reads, saves and exports exclusively in open standard formats such as FASTA, GenBank, EMBL, or UniProt; the same formats that you can open and read with a simple text editor:
Advanced Sequence Viewer
The viewer offer a detailed representation of the sequence and its features with a choice of varied ladders:
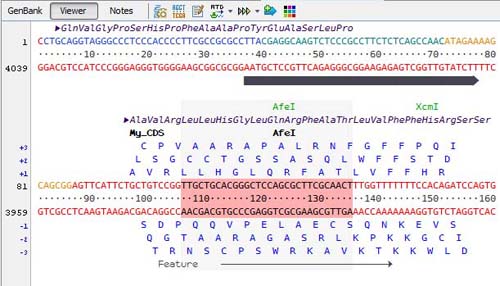
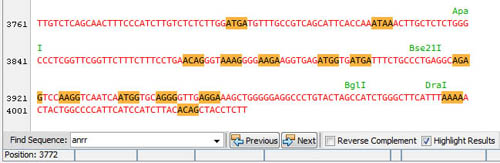
Restriction enzyme sites are indicated on a separate line and when selected the corresponding sites are highlighted on the sequence:
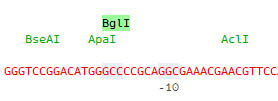
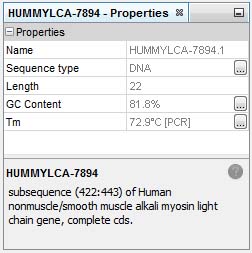
Detailed information is immediatly available in the Properties window:
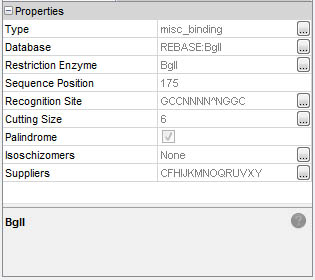
Select a feature by simple click:
Double-click the feature whilte pressing the Control key to select the sequence:
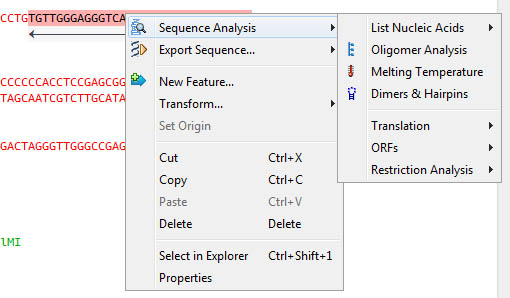
Right-click to acces sequence analysis, copy to clipboard, export in varied formats:
Information of sequence selection is immediatly available in the status bar:

For short sequence <100 bp %GC content and melting temperature for PCR are shown in the Properties window (see Simple and Rapid Primer Design).
Advanced Search Capabilities
Search as you type in the sequence viewer, iterate through the results, search the reverse complement sequence as you wish:
Advanced Restriction Analysis
Select, filter retriction enzymes and conduct varied analyses from the Restriction Enzymes window:
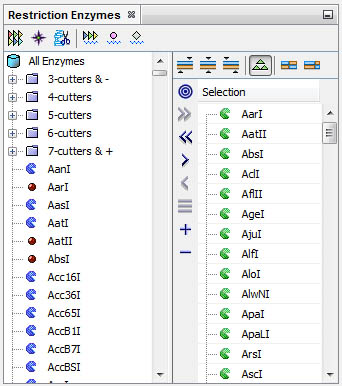
You can select the set of enzymes cutting one sequence; filter for particular properties such as 6-cutters and cut a different sequence to find out common or exclusive enzymes.
Feature Navigator
Get a complete view of all sequence features in the Navigator window:
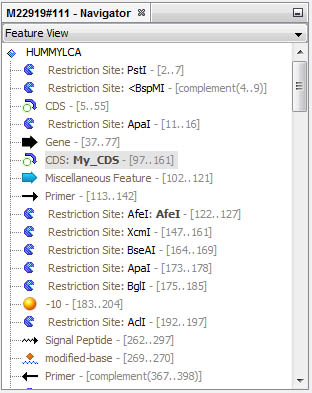
Features are automatically selected in the Navigator when you circulate in the viewer; inversely, you can select the corresponding sequence directly from the Navigator window.
Simple & Rapid Primer Design
All experience molecular biologists know that most primers will work as expected; what you want is being able to pinpoint the few that will waste a lot of your precious time. In GenBeans, you know %GC content and Tm for PCR as you select the sequence, so you can pick the primer with the proper annealing temperature as you go:
From the click of a button, you have access to a complete oligomer analysis,melting temperature from varied calculation methods or conditions, and detection of hairpins and dimers:
Instead of doing exhausting calculations to determine the complete secondary structure of your oligos, GenBeans uses a novel and fast algorythm to detect local sequences likely responsible for the formation of low energy, stable conformations. This includes base pairing, simple buldges and loops. Here is as an example of a well-hidden and quite stable dimer:

Access large sequence files
Increase the parsing limit to open large sequences. We loaded the entire IgH locus in less than 10 s in the viewer!
